Today marks an important milestone in the Nextflow project. We are thrilled to announce three important changes to better meet users’ needs and ground the project on a solid foundation upon which to build a vibrant ecosystem of tools and data analysis applications for genomic research and beyond.
Nextflow was originally licensed as GPLv3 open source software more than five years ago. GPL is designed to promote the adoption and spread of open source software and culture. On the other hand it has also some controversial side-effects, such as the one on derivative works and legal implications which make the use of GPL released software a headache in many organisations. We have previously discussed these concerns in this blog post and, after community feedback, have opted to change the project license to Apache 2.0.
This is a popular permissive free software license written by the Apache Software Foundation (ASF). Software distributed with this license requires the preservation of the copyright notice and disclaimer. It allows the freedom to use the software for any purpose, to distribute it, to modify it, and to distribute modified versions of the software without dictating the licence terms of the resulting applications and derivative works. We are sure this licensing model addresses the concerns raised by the Nextflow community and will boost further project developments.
In the time since Nextflow was open sourced, we have released 150 versions which have been used by many organizations to deploy critical production workflows on a large range of computational platforms and under heavy loads and stress conditions.
For example, at the Centre for Genomic Regulation (CRG) alone, Nextflow has been used to deploy data intensive computation workflows since 2014, and it has orchestrated the execution of over 12 million jobs totalling 1.4 million CPU-hours.
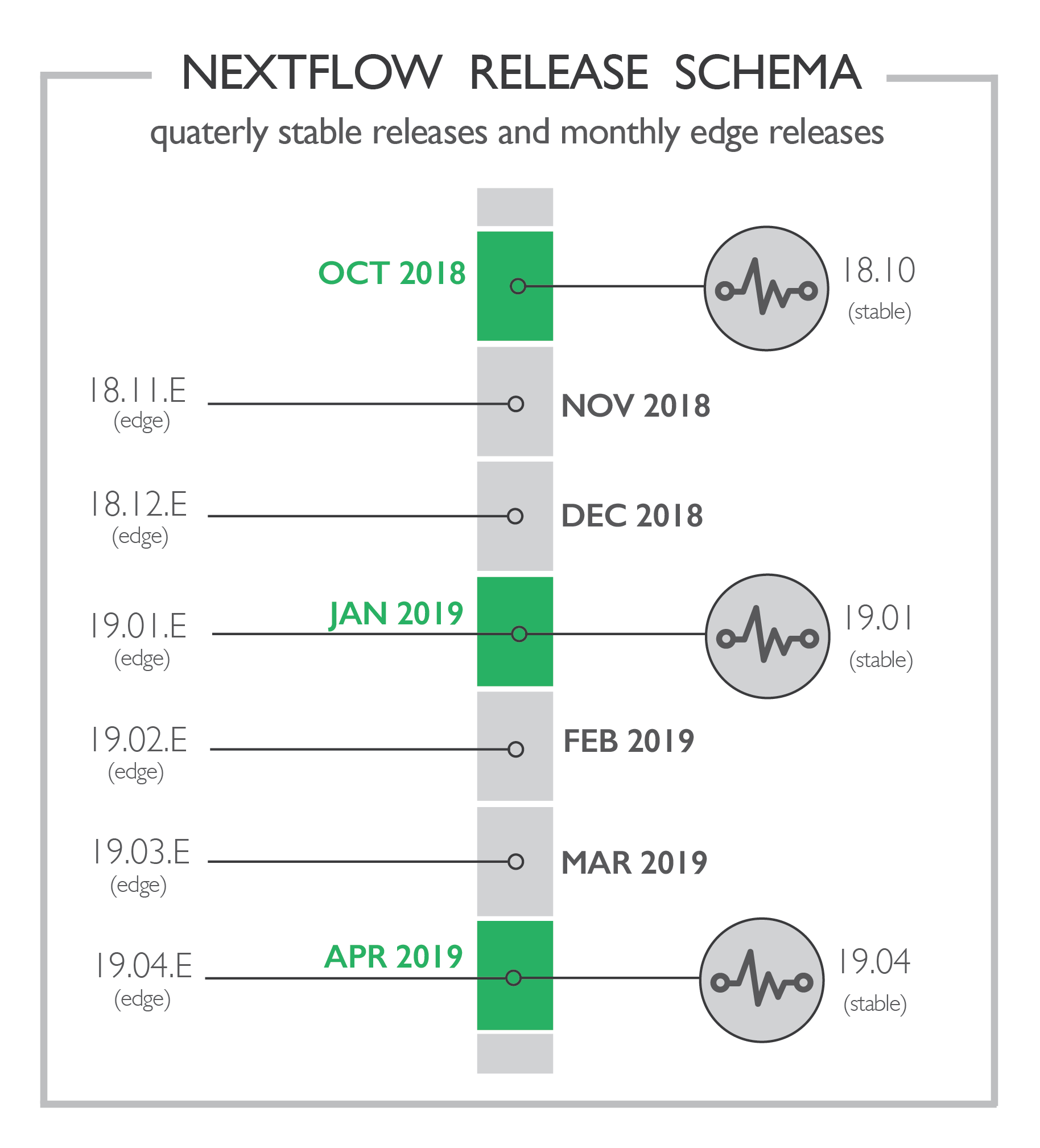
This extensive use across different execution environments has resulted in a reliable software package, and it’s therefore finally time to declare Nextflow stable and drop the zero from the version number!
From today onwards, Nextflow will use a 3 monthly time-based stable release cycle. Today’s release is numbered as 18.10, the next one will be on January 2019, numbered as 19.01, and so on. This gives our users a more predictable release cadence and allows us to better focus on new feature development and scheduling.
Along with the 3-months stable release cycle, we will provide a monthly edge release, which will include access to the latest experimental features and developments. As such, it should only be used for evaluation and testing purposes.
Finally, for organisations requiring commercial support, we have recently incorporated Seqera Labs, a spin-off of the Centre for Genomic Regulation.
Seqera Labs will foster Nextflow adoption as professional open source software by providing commercial support services and exploring new innovative products and solutions.
It’s important to highlight that Seqera Labs will not close or make Nextflow a commercial project. Nextflow is and will continue to be owned by the CRG and the other contributing organisations and individuals.
The Nextflow project has reached an important milestone. In the last five years it has grown and managed to become a stable technology used by thousands of people daily to deploy large scale workloads for life science data analysis applications and beyond. The project is now exiting from the experimental stage.
With the above changes we want to fulfil the needs of researchers, for a reliable tool enabling scalable and reproducible data analysis, along with the demand of production oriented users, who require reliable support and services for critical deployments.
Above all, our aim is to strengthen the community effort around the Nextflow ecosystem and make it a sustainable and solid technology in the long run.
We want to say thank you to all the people who have supported and contributed to this project to this stage. First of all to Cedric Notredame for his long term commitment to the project within the Comparative Bioinformatics group at CRG. The Open Bioinformatics Foundation (OBF) in the name of Chris Fields and The Ontario Institute for Cancer Research (OICR), namely Dr Lincoln Stein, for supporting the Nextflow change of license. The CRG TBDO department, and in particular Salvatore Cappadona for his continued support and advice. Finally, the user community who with their feedback and constructive criticism contribute everyday to make this project more stable, useful and powerful.